|
I am an AI/ML Researcher at Apple. My research focuses on machine learning and multimodal learning. Presently, I work on controllable generative models for 3D computer vision and speech/audio. Previously, I was a Ph.D. student at the Laboratory for Information & Decision Systems at MIT working with Caroline Uhler. During my Ph.D., I interned at Apple, Niantic Labs, Meta Reality Labs, Bosch Center for AI, and Adobe Research. Email / CV / Google Scholar |

|
|
Research Topics: all / multimodal learning / audio-visual learning / computational biology / generative modeling / optimal transport / causal inference |
|
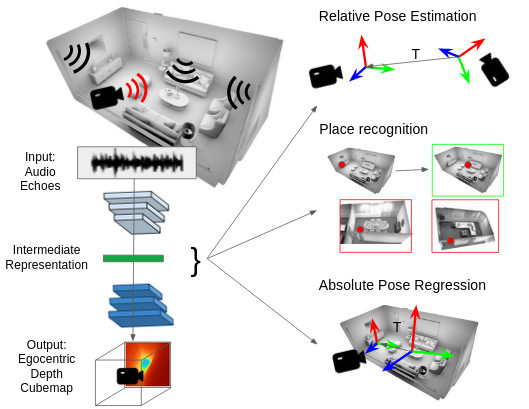
Karren Yang, Clément Godard, Eric Brachmann, Michael Firman ECCV, 2022 We use audio sensing to improve the performance of visual localization methods on three tasks: relative pose estimation, place recognition, and absolute pose regression. pdf | abstractIn this work, we show how to estimate a device's position and orientation indoors by echolocation, i.e., by interpreting the echoes of an audio signal that the device itself emits. Established visual localization methods rely on the device's camera and yield excellent accuracy if unique visual features are in view and depicted clearly. We argue that audio sensing can offer complementary information to vision for device localization, since audio is invariant to adverse visual conditions and can reveal scene information beyond a camera's field of view. We first propose a strategy for learning an audio representation that captures the scene geometry around a device using supervision transfer from vision. Subsequently, we leverage this audio representation to complement vision in three device localization tasks: relative pose estimation, place recognition, and absolute pose regression. Our proposed methods outperform state-of-the-art vision models on new audio-visual benchmarks for the Replica and Matterport3D datasets. |
|
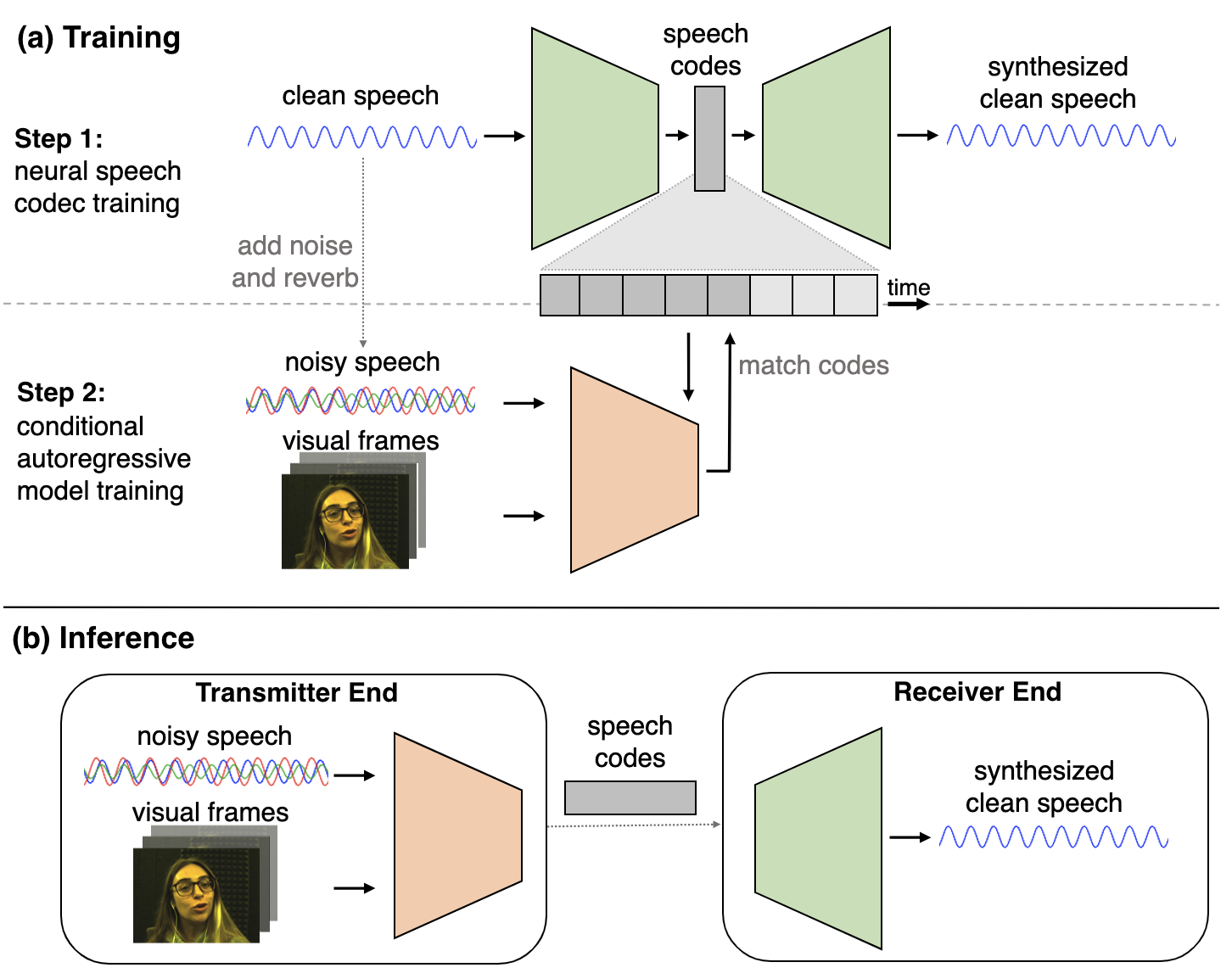
Karren Yang, Dejan Markovic, Steven Krenn, Vasu Agrawal, Alexander Richard CVPR, 2022 (Oral Presentation) We perform visual speech enhancement by using audio-visual speech cues to generate the codes of a neural speech codec, enabling efficient synthesis of clean, realistic speech from noisy signals. pdf | abstract | video | datasetSince facial actions such as lip movements contain significant information about speech content, it is not surprising that audio-visual speech enhancement methods are more accurate than their audio-only counterparts. Yet, state-of-the-art approaches still struggle to generate clean, realistic speech without noise artifacts and unnatural distortions in challenging acoustic environments. In this paper, we propose a novel audio-visual speech enhancement framework for high-fidelity telecommunications in AR/VR. Our approach leverages audio-visual speech cues to generate the codes of a neural speech codec, enabling efficient synthesis of clean, realistic speech from noisy signals. Given the importance of speaker-specific cues in speech, we focus on developing personalized models that work well for individual speakers. We demonstrate the efficacy of our approach on a new audio-visual speech dataset collected in an unconstrained, large vocabulary setting, as well as existing audio-visual datasets, outperforming speech enhancement baselines on both quantitative metrics and human evaluation studies. Please see the supplemental video for qualitative results. |
|
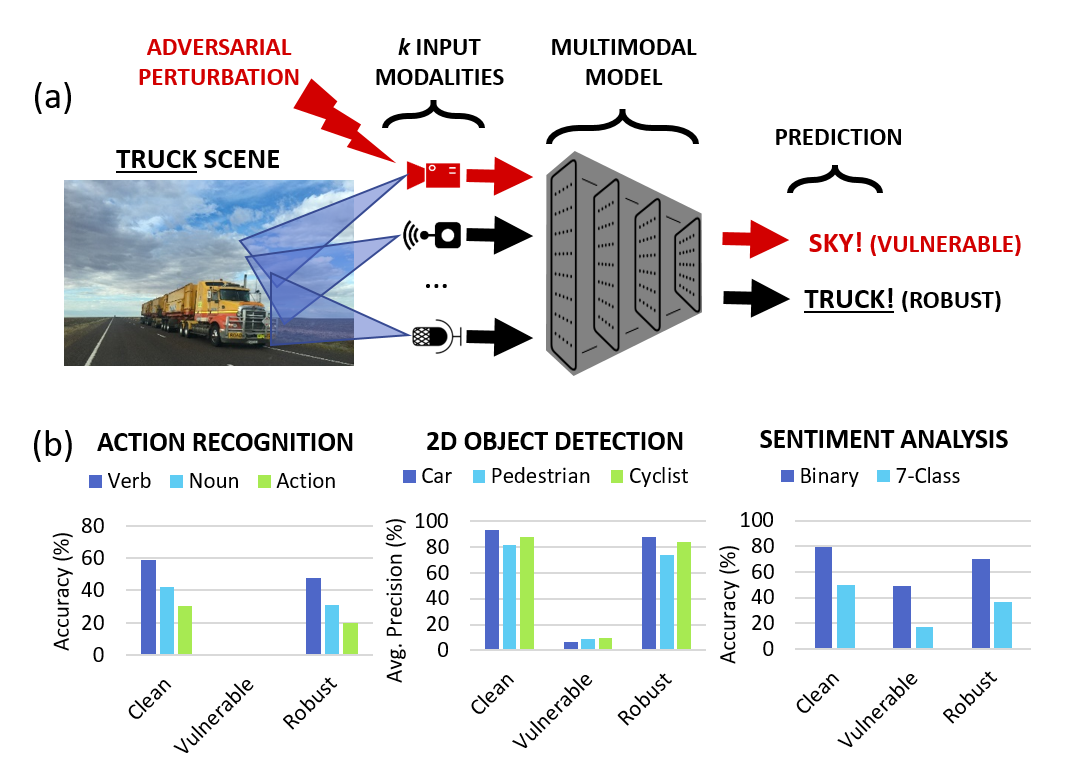
Karren Yang, Wan-Yi Lin, Manash Barman, Filipe Condessa, Zico Kolter CVPR, 2021 We study the robustness of multimodal models on three tasks-- action recognition, object detection, and sentiment analysis-- and develop a robust fusion strategy that protects against worst-case errors caused by a single modality. pdf | abstractBeyond achieving high performance across many vision tasks, multimodal models are expected to be robust to single-source faults due to the availability of redundant information between modalities. In this paper, we investigate the robustness of multimodal neural networks against worst-case (i.e., adversarial) perturbations on a single modality. We first show that standard multimodal fusion models are vulnerable to single-source adversaries: an attack on any single modality can overcome the correct information from multiple unperturbed modalities and cause the model to fail. This surprising vulnerability holds across diverse multimodal tasks and necessitates a solution. Motivated by this finding, we propose an adversarially robust fusion strategy that trains the model to compare information coming from all the input sources, detect inconsistencies in the perturbed modality compared to the other modalities, and only allow information from the unperturbed modalities to pass through. Our approach significantly improves on state-of-the-art methods in single-source robustness, achieving gains of 7.8-25.2% on action recognition, 19.7-48.2% on object detection, and 1.6-6.7% on sentiment analysis, without degrading performance on unperturbed (i.e., clean) data. |
|
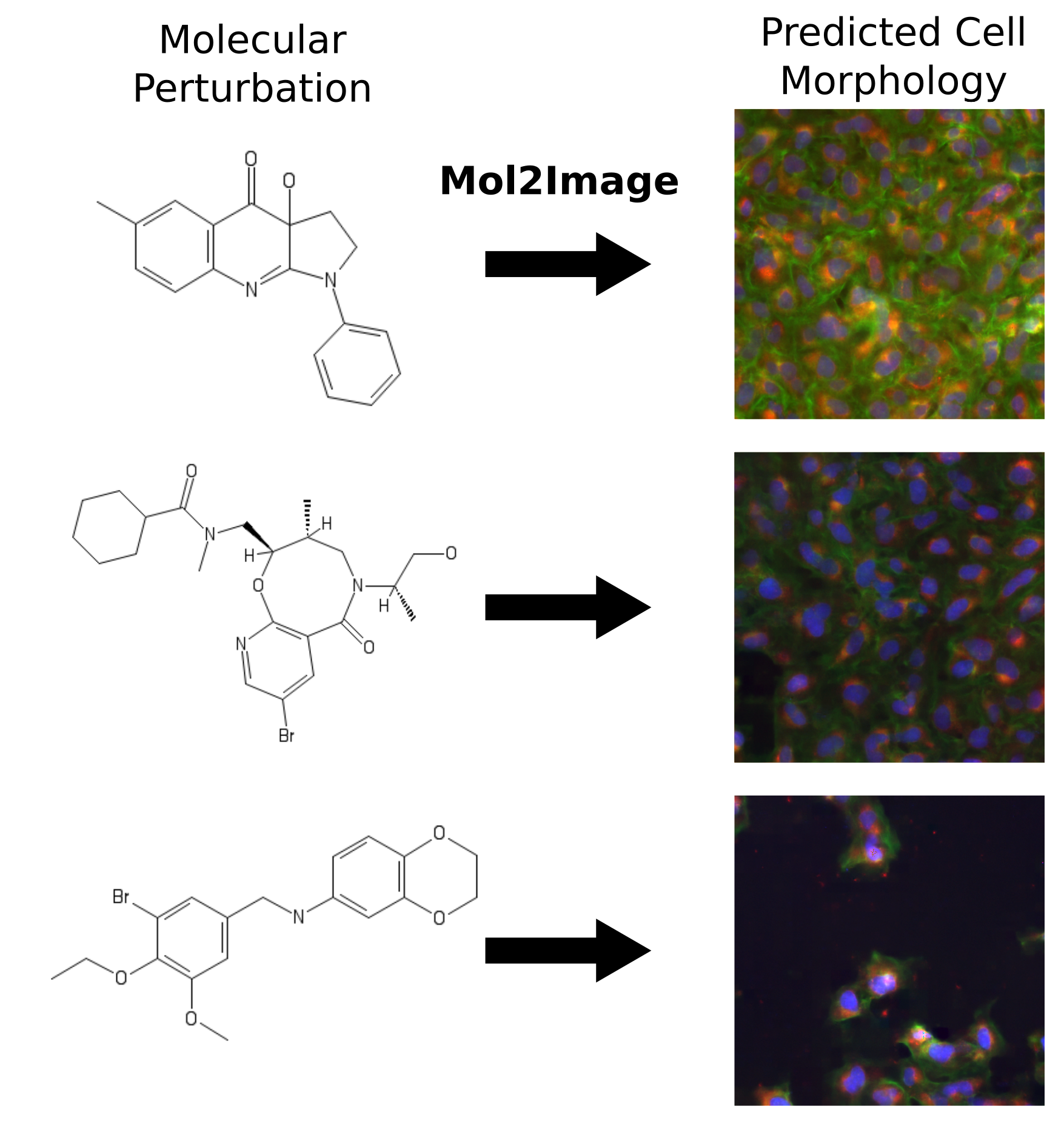
Karren Yang, Samuel Goldman, Wengong Jin, Alex Lu, Regina Barzilay, Tommi Jaakkola, Caroline Uhler CVPR, 2021 We build a molecule-to-image synthesis model that predicts the biological effects of molecular treatments on cell microscopy images. pdf | abstract | codeIn this paper, we aim to synthesize cell microscopy images under different molecular interventions, motivated by practical applications to drug development. Building on the recent success of graph neural networks for learning molecular embeddings and flow-based models for image generation, we propose Mol2Image: a flow-based generative model for molecule to cell image synthesis. To generate cell features at different resolutions and scale to high-resolution images, we develop a novel multi-scale flow architecture based on a Haar wavelet image pyramid. To maximize the mutual information between the generated images and the molecular interventions, we devise a training strategy based on contrastive learning. To evaluate our model, we propose a new set of metrics for biological image generation that are robust, interpretable, and relevant to practitioners. We show quantitatively that our method learns a meaningful embedding of the molecular intervention, which is translated into an image representation reflecting the biological effects of the intervention. |
|
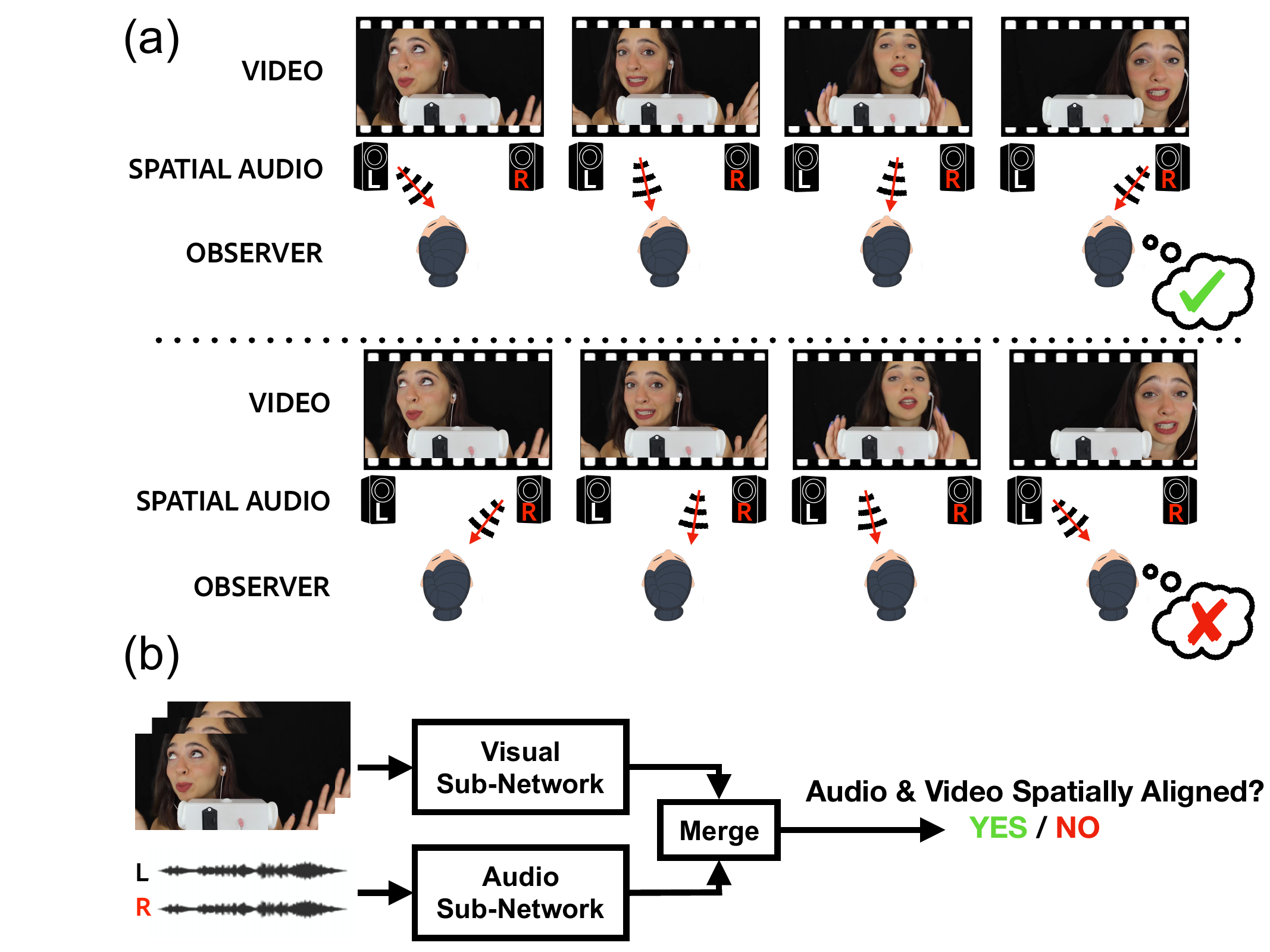
Karren Yang, Justin Salamon, Bryan Russell CVPR, 2020 (Oral Presentation) We leverage spatial correspondence between audio and vision in videos for self-supervised representation learning and apply the learned representations to three downstream tasks: sound localization, audio spatialization, and audio-visual sound separation. pdf | abstract | project website | dataset | demo codeSelf-supervised audio-visual learning aims to capture useful representations of video by leveraging correspondences between visual and audio inputs. Existing approaches have focused primarily on matching semantic information between the sensory streams. We propose a novel self-supervised task to leverage an orthogonal principle: matching spatial information in the audio stream to the positions of sound sources in the visual stream. Our approach is simple yet effective. We train a model to determine whether the left and right audio channels have been flipped, forcing it to reason about spatial localization across the visual and audio streams. To train and evaluate our model, we introduce a large-scale video dataset, YouTube-ASMR-300K, with spatial audio comprising over 900 hours of footage. We demonstrate that understanding spatial correspondence enables models to perform better on three audio-visual tasks, achieving quantitative gains over supervised and self-supervised baselines that do not leverage spatial audio cues. We also show how to extend our self-supervised approach to 360 degree videos with ambisonic audio. |
|
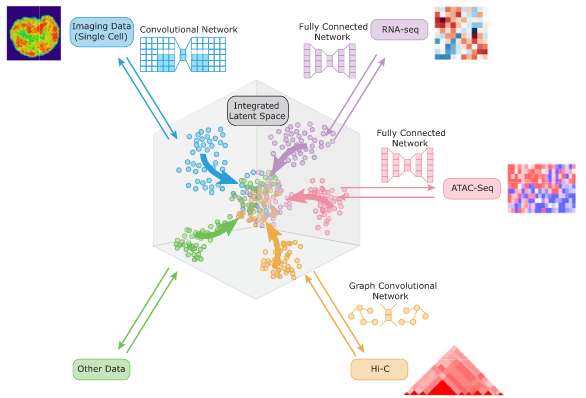
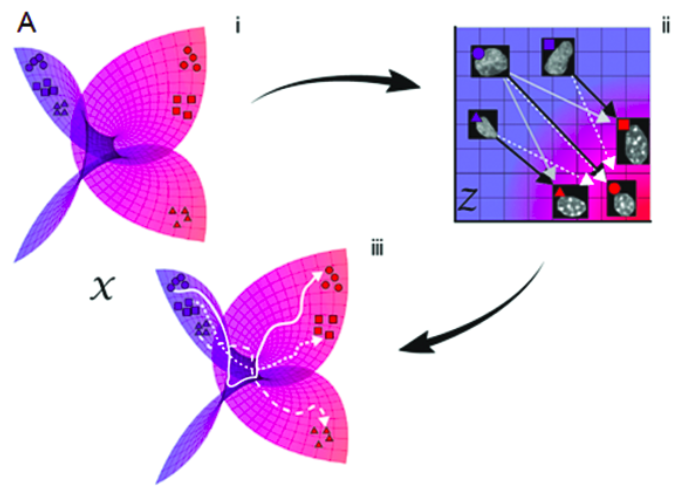
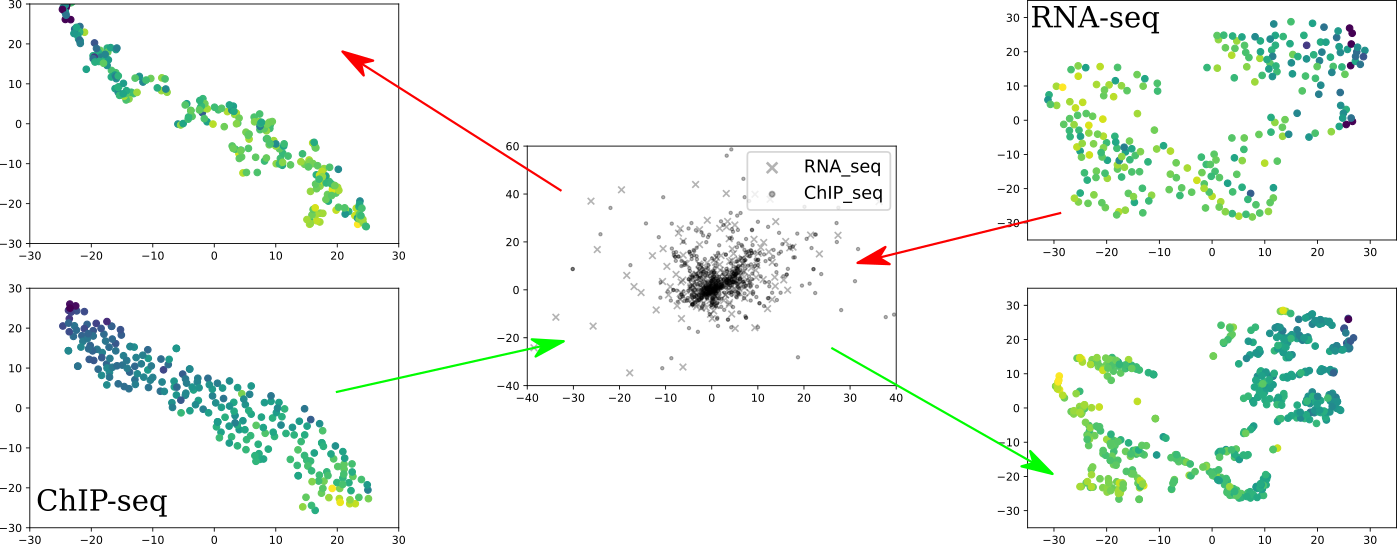
Karren Dai Yang*, Anastasiya Belyaeva*, Saradha Venkatachalapathy, Karthik Damodaran, Abigail Katcoff, Adityanarayanan Radhakrishnan, G. V. Shivashankar, Caroline Uhler Nature Communications 12, 31 (2021) We propose a framework for integrating and translating between different modalities of single-cell biological data by using autoencoders to map to a shared latent space. pdf | abstract | codeThe development of single-cell methods for capturing different data modalities including imaging and sequencing has revolutionized our ability to identify heterogeneous cell states. Different data modalities provide different perspectives on a population of cells, and their integration is critical for studying cellular heterogeneity and its function. While various methods have been proposed to integrate different sequencing data modalities, coupling imaging and sequencing has been an open challenge. We here present an approach for integrating vastly different modalities by learning a probabilistic coupling between the different data modalities using autoencoders to map to a shared latent space. We validate this approach by integrating single-cell RNA-seq and chromatin images to identify distinct subpopulations of human naive CD4+ T-cells that are poised for activation. Collectively, our approach provides a framework to integrate and translate between data modalities that cannot yet be measured within the same cell for diverse applications in biomedical discovery. |
|
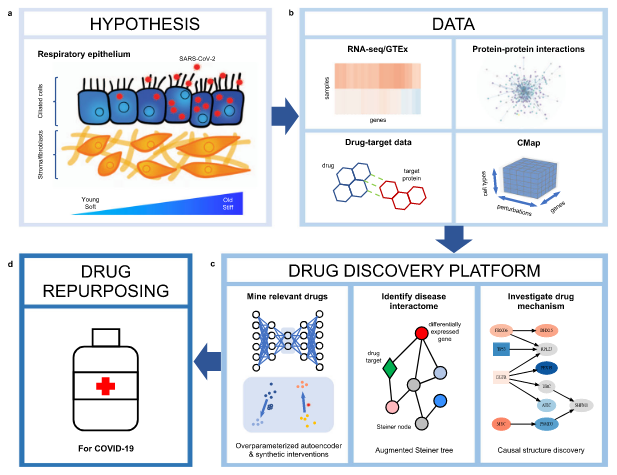
Anastasiya Belyaeva, Louis Cammarata, Adityanarayanan Radhakrishnan, Chandler Squires, Karren Yang, G. V. Shivashankar, Caroline Uhler Nature Communications 12, 1024 (2021) We integrate transcriptomic, proteomic, and structural data to identify candidate drugs and targets that affect the SARS-CoV-2 and aging pathways. pdf | abstractGiven the severity of the SARS-CoV-2 pandemic, a major challenge is to rapidly repurpose existing approved drugs for clinical interventions. While a number of data-driven and experimental approaches have been suggested in the context of drug repurposing, a platform that systematically integrates available transcriptomic, proteomic and structural data is missing. More importantly, given that SARS-CoV-2 pathogenicity is highly age-dependent, it is critical to integrate aging signatures into drug discovery platforms. We here take advantage of large-scale transcriptional drug screens combined with RNA-seq data of the lung epithelium with SARS-CoV-2 infection as well as the aging lung. To identify robust druggable protein targets, we propose a principled causal framework that makes use of multiple data modalities. Our analysis highlights the importance of serine/threonine and tyrosine kinases as potential targets that intersect the SARS-CoV-2 and aging pathways. By integrating transcriptomic, proteomic and structural data that is available for many diseases, our drug discovery platform is broadly applicable. Rigorous in vitro experiments as well as clinical trials are needed to validate the identified candidate drugs. |
|
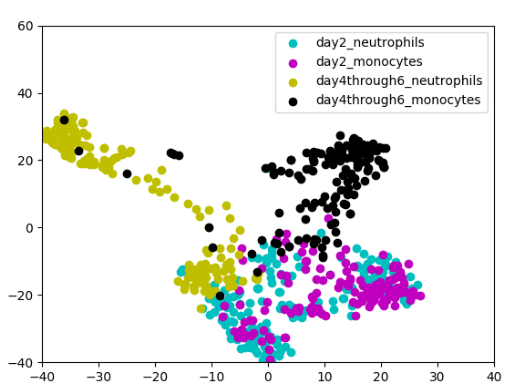
Neha Prasad, Karren Yang, Caroline Uhler ICML Workshop on Computational Biology, 2020 (Oral Spotlight) We propose a novel approach to computational lineage tracing that combines supervised learning with optimal transport based on generative adversarial networks. pdf | abstract | codeIn this paper, we present Super-OT, a novel approach to computational lineage tracing that combines a supervised learning framework with optimal transport based on Generative Adversarial Networks (GANs). Unlike previous approaches to lineage tracing, Super-OT has the flexibility to integrate paired data. We benchmark Super-OT based on single-cell RNA-seq data against Waddington-OT, a popular approach for lineage tracing that also employs optimal transport. We show that Super-OT achieves gains over Waddington-OT in predicting the class outcome of cells during differentiation, since it allows the integration of additional information during training. |

|
Karren Yang, Caroline Uhler ICLR, 2019 We align and translate between datasets by performing unbalanced optimal transport with generative adversarial networks. pdf | abstract | codeGenerative adversarial networks (GANs) are an expressive class of neural generative models with tremendous success in modeling high-dimensional continuous measures. In this paper, we present a scalable method for unbalanced optimal transport (OT) based on the generative-adversarial framework. We formulate unbalanced OT as a problem of simultaneously learning a transport map and a scaling factor that push a source measure to a target measure in a cost-optimal manner. In addition, we propose an algorithm for solving this problem based on stochastic alternating gradient updates, similar in practice to GANs. We also provide theoretical justification for this formulation, showing that it is closely related to an existing static formulation by Liero et al. (2018), and perform numerical experiments demonstrating how this methodology can be applied to population modeling. |
|
Karren Yang, Karthik Damodaran, Saradha Venkatachalapathy, Ali C. Soylemezoglu, G. V. Shivashankar, Caroline Uhler PLOS Computational Biology 16(4): e1007828 (2020) We combine autoencoding and optimal transport to align biological imaging datasets collected from different time points. pdf | abstract | codeLineage tracing involves the identification of all ancestors and descendants of a given cell, and is an important tool for studying biological processes such as development and disease progression. However, in many settings, controlled time-course experiments are not feasible, for example when working with tissue samples from patients. Here we present ImageAEOT, a computational pipeline based on autoencoders and optimal transport for predicting the lineages of cells using time-labeled datasets from different stages of a cellular process. Given a single-cell image from one of the stages, ImageAEOT generates an artificial lineage of this cell based on the population characteristics of the other stages. These lineages can be used to connect subpopulations of cells through the different stages and identify image-based features and biomarkers underlying the biological process. To validate our method, we apply ImageAEOT to a benchmark task based on nuclear and chromatin images during the activation of fibroblasts by tumor cells in engineered 3D tissues. We further validate ImageAEOT on chromatin images of various breast cancer cell lines and human tissue samples, thereby linking alterations in chromatin condensation patterns to different stages of tumor progression. Our results demonstrate the promise of computational methods based on autoencoding and optimal transport principles for lineage tracing in settings where existing experimental strategies cannot be used. |
|
Karren Yang, Caroline Uhler ICML Workshop on Computational Biology, 2019 (Oral Spotlight) We train domain-specific autoencoders to map different data modalities to the same latent space and translate between them. pdf | abstractMulti-domain translation seeks to learn a probabilistic coupling between marginal distributions that reflects the correspondence between different domains. We assume that data from different domains are generated from a shared latent representation based on a structural equation model. Under this assumption, we show that the problem of computing a probabilistic coupling between marginals is equivalent to learning multiple uncoupled autoencoders that embed to a given shared latent distribution. In addition, we propose a new framework and algorithm for multi-domain translation based on learning the shared latent distribution and training autoencoders under distributional constraints. A key practical advantage of our framework is that new autoencoders (i.e., new domains) can be added sequentially to the model without retraining on the other domains, which we demonstrate experimentally on image as well as genomics datasets. |
|
Raj Agrawal, Chandler Squires, Karren Yang, Karthik Shanmugam, Caroline Uhler AISTATS, 2019 We propose an experimental design strategy for target causal discovery. pdf | abstract | codeDetermining the causal structure of a set of variables is critical for both scientific inquiry and decision-making. However, this is often challenging in practice due to limited interventional data. Given that randomized experiments are usually expensive to perform, we propose a general framework and theory based on optimal Bayesian experimental design to select experiments for targeted causal discovery. That is, we assume the experimenter is interested in learning some function of the unknown graph (e.g., all descendants of a target node) subject to design constraints such as limits on the number of samples and rounds of experimentation. While it is in general computationally intractable to select an optimal experimental design strategy, we provide a tractable implementation with provable guarantees on both approximation and optimization quality based on submodularity. We evaluate the efficacy of our proposed method on both synthetic and real datasets, thereby demonstrating that our method realizes considerable performance gains over baseline strategies such as random sampling. |
|
Karren Yang, Abigail Katcoff, Caroline Uhler ICML, 2018 (Oral Presentation) We characterize interventional Markov equivalence classes of DAGs that can be identified under soft interventions and propose the first provably consistent algorithm for learning DAGs in this setting. pdf | abstractWe consider the problem of learning causal DAGs in the setting where both observational and interventional data is available. This setting is common in biology, where gene regulatory networks can be intervened on using chemical reagents or gene deletions. Hauser and Bühlmann (2012) previously characterized the identifiability of causal DAGs under perfect interventions, which eliminate dependencies between targeted variables and their direct causes. In this paper, we extend these identifiability results to general interventions, which may modify the dependencies between targeted variables and their causes without eliminating them. We define and characterize the interventional Markov equivalence class that can be identified from general (not necessarily perfect) intervention experiments. We also propose the first provably consistent algorithm for learning DAGs in this setting and evaluate our algorithm on simulated and biological datasets. |

|
Adityanarayanan Radhakrishnan, Karren Yang, Mikhail Belkin, Caroline Uhler arXiv, 2018 We show that overparameterized autoencoders are biased towards learning step function around training examples. pdf | abstractThe ability of deep neural networks to generalize well in the overparameterized regime has become a subject of significant research interest. We show that overparameterized autoencoders exhibit memorization, a form of inductive bias that constrains the functions learned through the optimization process to concentrate around the training examples, although the network could in principle represent a much larger function class. In particular, we prove that single-layer fully-connected autoencoders project data onto the (nonlinear) span of the training examples. In addition, we show that deep fully-connected autoencoders learn a map that is locally contractive at the training examples, and hence iterating the autoencoder results in convergence to the training examples. Finally, we prove that depth is necessary and provide empirical evidence that it is also sufficient for memorization in convolutional autoencoders. Understanding this inductive bias may shed light on the generalization properties of overparametrized deep neural networks that are currently unexplained by classical statistical theory. |
|
Yuhao Wang, Liam Solus, Karren Yang, Caroline Uhler NeurIPS, 2017 (Oral Presentation) We present two provably consistent algorithms for learning DAGs from observational and (hard) interventional data. pdf | abstractLearning directed acyclic graphs using both observational and interventional data is now a fundamentally important problem due to recent technological developments in genomics that generate such single-cell gene expression data at a very large scale. In order to utilize this data for learning gene regulatory networks, efficient and reliable causal inference algorithms are needed that can make use of both observational and interventional data. In this paper, we present two algorithms of this type and prove that both are consistent under the faithfulness assumption. These algorithms are interventional adaptations of the Greedy SP algorithm and are the first algorithms using both observational and interventional data with consistency guarantees. Moreover, these algorithms have the advantage that they are nonparametric, which makes them useful also for analyzing non-Gaussian data. In this paper, we present these two algorithms and their consistency guarantees, and we analyze their performance on simulated data, protein signaling data, and single-cell gene expression data. |
|
|